Protein-arginine deiminase type-2 (Mus musculus)
| Basic Information | |
|---|---|
| Short Name | PADI2, PDI2 |
| UNP ID | Q9ULC6 |
| Organism | Mus musculus |
| Cys Site | Cys655 |
| Family/Domain | Protein arginine deiminase family |
| Known Ligand | Ligand list |
| Function Type | Metabolic enzyme |
Summary
Protein Function
Protein-arginine deiminase type-2 is a member of the peptidyl arginine deiminase family of enzymes, which catalyze the post-translational deimination of proteins by converting arginine residues into citrullines in the presence of calcium ions. The family members have distinct substrate specificities and tissue-specific expression patterns. The type II enzyme is the most widely expressed family member. Known substrates for this enzyme include myelin basic protein in the central nervous system and vimentin in skeletal muscle and macrophages. This enzyme is thought to play a role in the onset and progression of neurodegenerative human disorders, including Alzheimer disease and multiple sclerosis, and it has also been implicated in glaucoma pathogenesis. This gene exists in a cluster with four other paralogous genes. (From Wikipedia)
Cys Function & Property
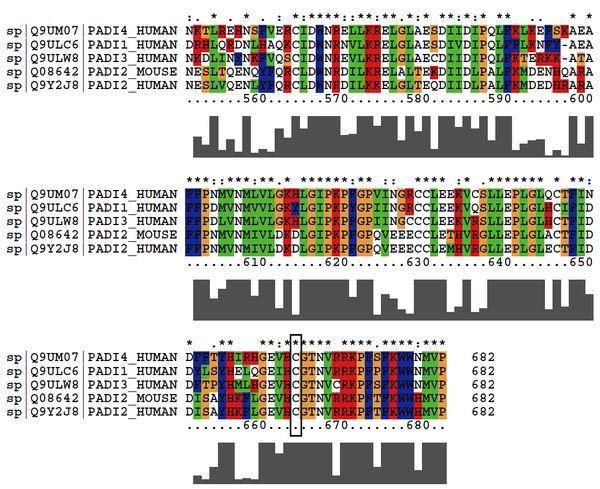
Cys655 is the active site of mouse PADI2. And this site is quite conserved in PADIs.
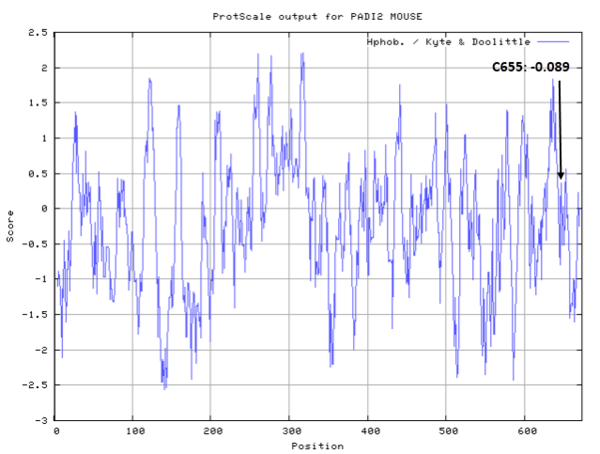
- Hydrophobic property:
- SASA:
- Cys655: Unknown
Protein Sequence
MQPPIRENML RERTVRLQYG SRVEAVYVLG TQLWTDVYSA APAGAKTFSL
KHSEGVKVEV VRDGEAEEVV TNGKQRWALS PSSTLRLSMA QASTEASSDK
VTVNYYEEEG SAPIDQAGLF LTAIEISLDV DADRDGEVEK NNPKKASWTW
GPEGQGAILL VNCDRDTPWL PKEDCSDEKV YSKQDLQDMS QMILRTKGPD
RLPAGYEIVL YISMSDSDKV GVFYVENPFF GQRYIHILGR QKLYHVVKYT
GGSAELLFFV EGLCFPDESF SGLVSIHVSL LEYMAEGIPL TPIFTDTVMF
RIAPWIMTPN ILPPVSVFVC CMKDNYLFLK EVKNLVEKTN CELKVCFQYM
NRGDRWIQDE IEFGYIEAPH KGFPVVLDSP RDGNLKDFPI KQLLGPDFGY
VTREPLFETV TSLDSFGNLE VSPPVTVNGK EYPLGRILIG SSFPLSGGRR
MTKVVRDFLQ AQQVQAPVEL YSDWLTVGHV DEFMTFIPIP GKKEFRLLMA
STSACYQLFR EKQKAGHGEA VMFKGLGGMS SKRITINKIL SNESLTQENQ
YFQRCLDWNR DILKRELALT EKDIIDLPAL FKMDENHQAR AFFPNMVNMI
VLDKDLGIPK PFGPQVEEEC CLETHVRGLL EPLGLACTFI DDISAYHKFL
GEVHCGTNVR RKPFTFKWWH MVP
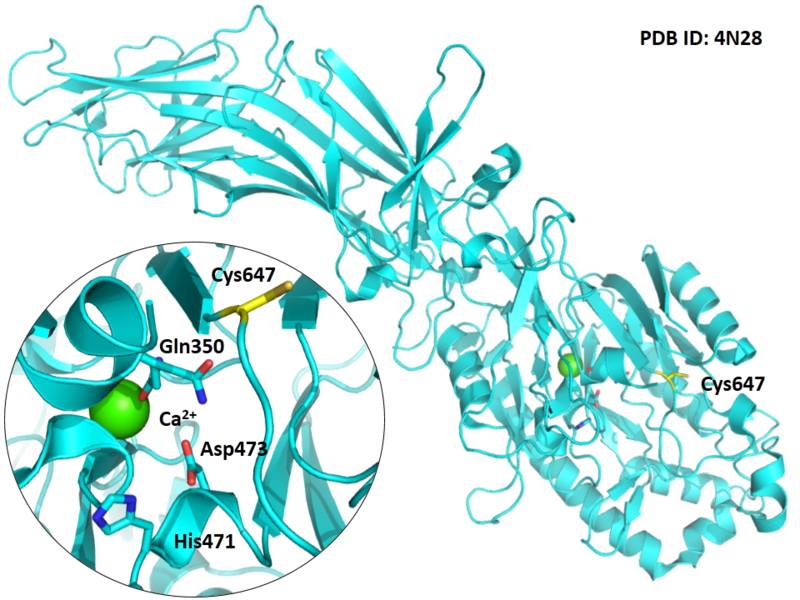
Structural Information
- Known structure with covalent ligand:
- Unknown
- Protein structure:
Related Pathway
- Unknown
Experimental Evidence
- Homologous Analysis Of Sequence, Molecular Docking
Reference
- Bello A M, Wasilewski E, Wei L, et al. Interrogation of the active sites of protein arginine deiminases (PAD1,-2, and-4) using designer probes[J]. ACS medicinal chemistry letters, 2013, 4(2): 249-253. 24900657